Screening
A Unique Screening Platform
A unique screening platform offering innovative solutions for a wide spectrum of targets.
Our unique phage display-screening platform allows us to quickly identify Bicycle® molecules specific for potential high value therapeutic targets. The platform has been optimised over many years to be efficient, allowing us to tune the development of these Bicycle molecules to precisely address each specific therapeutic application.
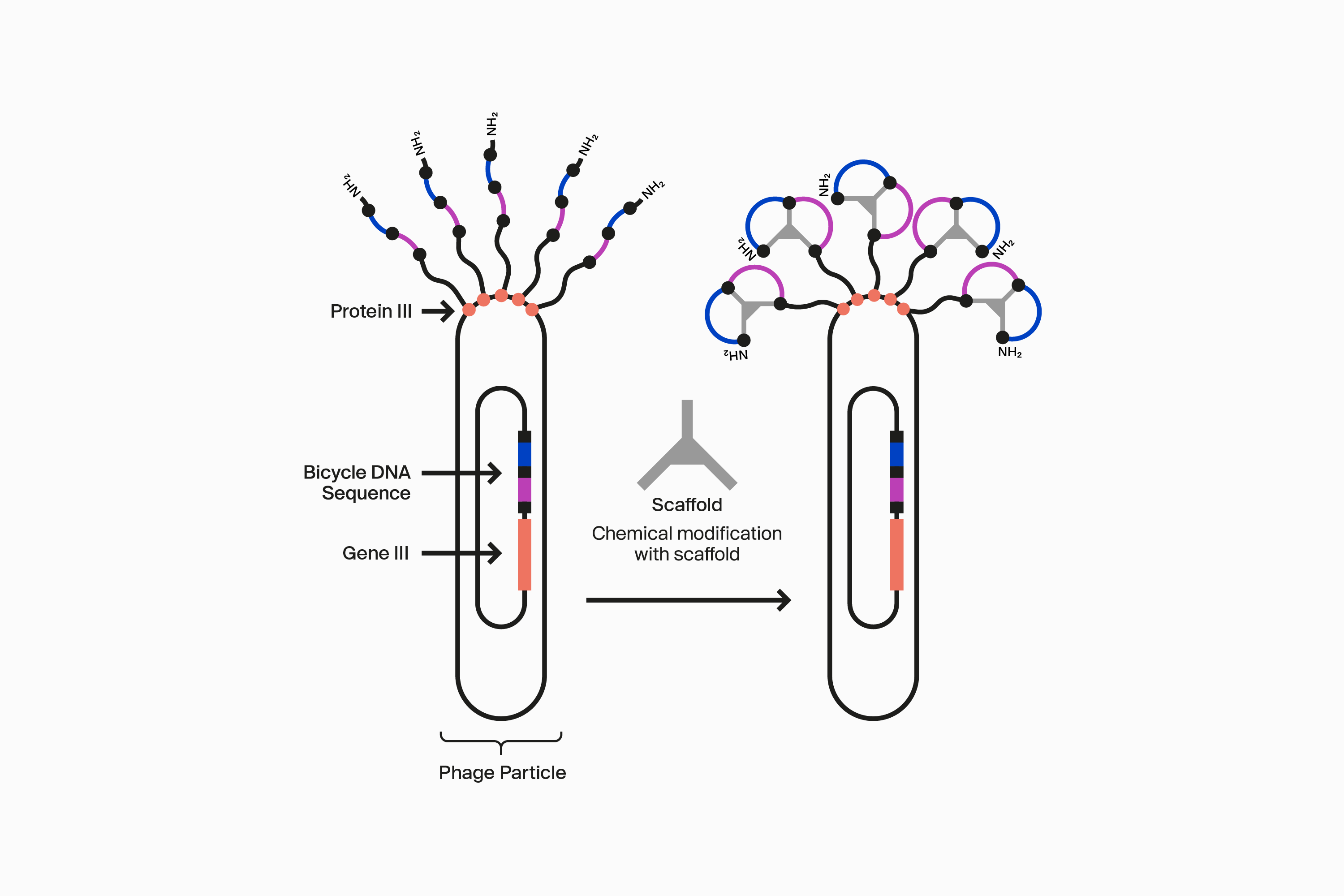
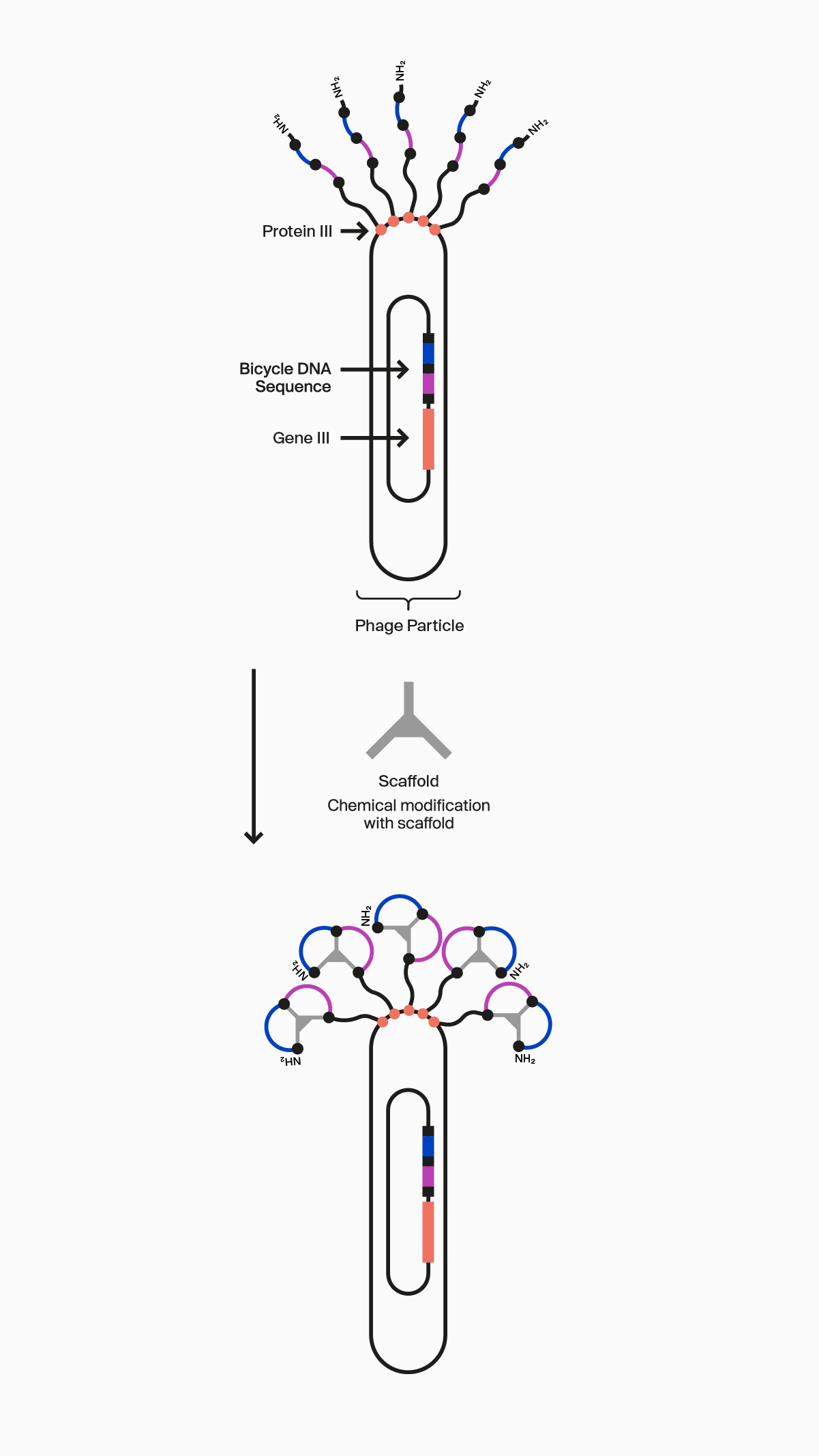
Our novel, proprietary phage display-screening platform uses synthetic biology to display a vast diversity (>10 20) of initially linear peptides on the surface of engineered bacteriophage (phage). These linear peptides are then covalently reacted to one of our many scaffolds through thioether bonds to strategically located cysteine residues in the peptide to form the Bicycle molecule structure on the surface of the phage particle. This incredible diversity is generated by changing the design of the Bicycle molecule at four key areas: firstly, each amino acid can be varied at each position between the cysteines within the rings; secondly, the number of amino acids can be changed in each ring; thirdly the symmetry of the rings can be altered; and finally, the choice of scaffold used changes the way the appended amino acids are presented. Collectively, this allows us to choose from over 300 different library configurations, enabling high chemical diversity and increasing the probability of successfully identifying target binders from our screening campaigns.
Bicycle molecules can be readily identified for a wide spectrum of diverse protein targets, including potentially those for which conventional small molecules have not proven successful. Our screening platform can be deployed to screen either soluble proteins or cell-based targets. In addition to being resource-efficient and rapid, the process uniquely uses an integral on-phage binding assay that informs Structure Activity Relationships (SAR). Once we have identified initial Bicycle molecules that bind the target, we next perform iterative rounds of affinity maturation to create chemical diversity around the common pharmacophore and then use the speed of the phage system to enrich and inform on SAR which provides invaluable information for our chemistry teams and the next stage of development (our chemical optimization phase). Here, we will make the Bicycle molecules synthetically on peptide synthesizers and replace the natural amino acids within the Bicycle molecule with non-natural ones to either improve affinity to the target or to add desirable drug-like properties, tuning the molecule with the aim that it will perform the exact function we need.
The final stage in the platform is to then put together the Bicycle therapeutic candidate for further development. As the screening process self-selects for Bicycle molecules that are payload-enabled and amenable to conjugation, this often involves us adding on other payloads such as cytotoxins, immune activators, nucleic acids or simply other Bicycle molecules. Bicycle molecules can be linked together with synthetic ease to create complex molecules with combinatorial pharmacology. Alternatively, Bicycle molecules in the form of multimers (where two or more Bicycle molecules, each binding a different target, are combined together) can also be used as standalone therapeutics, including those that we are exploring in our Bicycle® Tumor-Targeted Immune Cell Agonists (Bicycle TICAs) and Bicycle® NK-cell Tumor-Targeted Immune Cell Agonists (NK-TICAs) modulator programs. The flexibility of our Bicycle molecules, and our powerful screening platform, allows potential new therapeutic candidates to be rapidly conceived for further development across diverse therapeutic applications and a wide range of conditions.